JS research group
Gut microbiota and host-microbe interactions
RESEARCH

Our research focuses on gut microbiota interactions and host-microbe interactions. We aim to understand how bacterial communities of different complexities interact using systems biology approaches, and how their interactions affect host metabolism and metabolic diseases through their derived factors. We believe that the gut microbiome plays a central role in regulating host metabolic homeostasis. Our end goal is to develop next-generation precision microbiome therapies and intervention strategies to alleviate metabolic diseases and improve health.
Research 1. A general framework to study gut microbiota interactions.

Relevant publications: Sulaiman JE et al. Nature Communications, 15:7416 (2024).
Bottom-up construction of synthetic microbiomes combined with computational modeling and principled experimental design techniques can efficiently navigate large design landscapes of species combinations and allow us to reverse-engineer various ecological systems. We are using a bottom-up approach to decipher the ecological and molecular principles of gut microbiota interactions using a general framework that combines high-throughput experiments and computational modeling. We utilize the model to predict communities with optimized functions (e.g. pathogen inhibition or production of beneficial metabolites) and evaluate their functionality in mouse models.
Research 2. The impact of diet on pathogen-microbiota interactions.
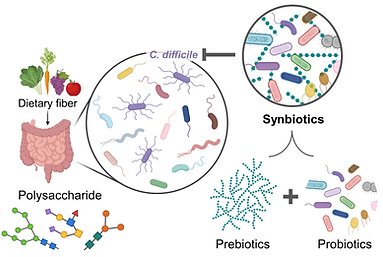
Relevant publications: Sulaiman JE et al. Frontiers in Microbiology, 16:1666747 (2025).
Consumption of dietary fiber significantly impacts the dynamics and functions of gut communities and has been associated with health benefits. Dietary fibers consist of polysaccharides, which are non-digestible forms of carbohydrates that are linked with glycosidic bonds. Not all gut bacteria can cleave these glycosidic linkages, and they rely on other microbes to break it down. Carbohydrate complexity in human diets has been found to limit microbial growth and reduce the sensitivity of human gut communities to perturbations (i.e. invasion with new species).
Nutrient environments have a huge impact on C. difficile colonization and growth. However, how dietary fibers impact inter-species interactions and colonization resistance of human gut microbiota to C. difficile remains unclear. We lack an understanding of how dietary fibers shape the interactions between C. difficile and human gut bacteria with different fiber-degrading capabilities. Our lab is interested in mapping inter-species interactions between C. difficile and human gut bacteria in different dietary fibers that resemble healthy human diets to inform dietary and bacterial interventions for C. difficile treatment. In addition, we are using the models trained on community data in different dietary fibers to design synbiotics that are effective in inhibiting C. difficile.
Research 3. Long-term growth experiments of pathogens in microbial communities.

Relevant publications: Sulaiman JE et al. Cell Host & Microbe, 33:42-58 (2025).
Bacterial pathogens could persistently colonize the human gut for yearlong timescales. Persistent colonization increases the risk of infection, acts as a reservoir for infection (i.e. increases the rate of transmission), and could result in evolutionary adaptation which alters the metabolism and virulence of the pathogen.
Community interactions affect the evolutionary adaptation of a species, but the effect of human gut microbiota on the evolution of many pathogens (e.g. C. difficile inestinal pathogen, or P. gingivalis oral pathogen) is still largely unknown. Our lab uses laboratory experiments involving the building of microbial communities to study the long-term growth behavior of pathogens with human gut bacteria and how communities influence pathogens’ adaptation. We use whole-genome sequencing and omics approaches to generate insights into their mechanisms of adaptation.
Research 4. Role of gut microbiota-derived metabolites on host metabolism and vice versa.

Relevant publications: Long K et al. Journal of Clinical Investigation,135:22 (2025).
Studies have shown that gut microbiota regulate adipose tissue expansion and lipid metabolism, promote browning of WAT and activity of BAT, and play essential roles in adipose tissue inflammation in obesity and related disorders. On the contrary, adipose tissue also controls the gut microbiota via its derived factors. Our lab will use animal models and human samples to study the cross-talk between gut microbiota and adipose tissues in metabolic diseases.
To get mechanistic insights on the molecular level, we are interested in exploring how microbiota-derived metabolites impact host metabolism and vice versa. For instance, bacteria convert dietary fibers to short-chain fatty acids (SCFAs), produce branched-chain amino acids (BCAAs), and modify primary to secondary bile acids, which impact host tissues and health. Adipose tissue also secretes adipokines that affect the functioning and balance of the gut microbiota. Through a deeper understanding of this complex relationship, our goal is to come up with a novel approach using defined microbiome therapeutics to improve host metabolism and alleviate/cure metabolic diseases.